Introduction
Yukawa's Laboratory (Plant Molecular Biology), Nagoya City University
Our group has been involved in developing an understanding of the mechanistic aspects of genes expression during transcription as well as translation in higher plants. Our overall aim is to identify the molecular components involved in these fundamental processes of gene expression and understand their regulation at molecular level. Development of in vitro systems that could transcribe any gene or translate any mRNA with high fidelity was central to achieve the above stated objectives. During the years, we have developed an in vitro transcription system from tobacco nuclei and in vitro translation system from tobacco chloroplasts, which are two major assets of our group and are central to most of our research activities. We hope that by developing a deeper understanding of these fundamental processes of life, our studies would have applications in solving global issues like, food, energy and climate change.
Research
Plant in vitro transcription system
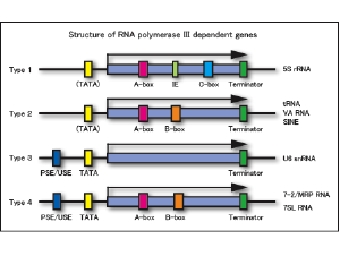
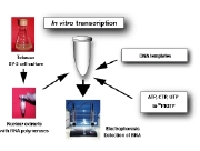 Schematic representation of in vitro transcripiton systemWe are studying various aspects of transcriptional regulation of nuclear genes under cell-free conditions. The in vitro system was developed from BY-2 tobacco cultured cells that are also known as HeLa cells of plants. As eukaryotic cells possess three different RNA polymerases, Pol I, Pol II and Pol III, our in vitro transcription system also supports all these activities. Since our knowledge of Pol III transcription is not as good as for the other two polymerases, our group’s recent efforts are concentrated towards understanding various aspects of Pol III activity. As show below, Pol III transcribed genes (class III genes) in plants have been classified into four types. This classification is different from that of animals and fungi. We therefore believe that a better understanding of plant Pol III based transcription would help in developing a comprehensive understanding of the eukaryotic transcription.
Schematic representation of in vitro transcripiton systemWe are studying various aspects of transcriptional regulation of nuclear genes under cell-free conditions. The in vitro system was developed from BY-2 tobacco cultured cells that are also known as HeLa cells of plants. As eukaryotic cells possess three different RNA polymerases, Pol I, Pol II and Pol III, our in vitro transcription system also supports all these activities. Since our knowledge of Pol III transcription is not as good as for the other two polymerases, our group’s recent efforts are concentrated towards understanding various aspects of Pol III activity. As show below, Pol III transcribed genes (class III genes) in plants have been classified into four types. This classification is different from that of animals and fungi. We therefore believe that a better understanding of plant Pol III based transcription would help in developing a comprehensive understanding of the eukaryotic transcription.
Chloroplast in vitro translation(Chloroplast Factory)
Chloroplast (left) and cytosolic (right) lysates including translation acitivityWe are studying various aspects of transcriptional regulation of nuclear genes under cell-free conditions. The in vitro system was developed from BY-2 tobacco cultured cells that are also known as HeLa cells of plants. As eukaryotic cells possess three different RNA polymerases, Pol I, Pol II and Pol III, our in vitro transcription system also supports all these activities. Since our knowledge of Pol III transcription is not as good as for the other two polymerases, our group’s recent efforts are concentrated towards understanding various aspects of Pol III activity. As show below, Pol III transcribed genes (class III genes) in plants have been classified into four types. This classification is different from that of animals and fungi. We therefore believe that a better understanding of plant Pol III based transcription would help in developing a comprehensive understanding of the eukaryotic transcription.
Identification of novel non-coding RNAs from higher plants
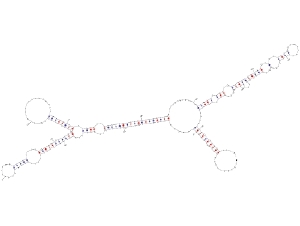 Predicted secondary structure of AtR8 RNAAfter completion of major projects on sequencing of plant genomes, annotation of various structural components of these genomes is in progress. According to the latest estimates the protein coding genes are not so many, however, a large number of non-protein-coding RNA (ncRNA) genes have been discovered. Although the protein coding genes can be predicted based on their structural features viz., open reading frame, and features of the upstream and downstream sequences, it is difficult to predict an ncRNA gene only from its sequence information. Because, there is no open reading frame and no sequence similarity, whatsoever so far found, in the characterized genes, in silico analysis has proven to be difficult. With an aim to discover novel ncRNA in higher plants, we are using the in vitro transcription system developed from tobacco cell nuclei. Our efforts have revealed some conserved regulatory elements in Pol III transcribed genes, which are being used to scan genomic data to find new ncRNA candidates. After their amplification by genomic PCR, these candidate genes can be validated for authenticity by using the in vitro transcription system developed in our lab.
Predicted secondary structure of AtR8 RNAAfter completion of major projects on sequencing of plant genomes, annotation of various structural components of these genomes is in progress. According to the latest estimates the protein coding genes are not so many, however, a large number of non-protein-coding RNA (ncRNA) genes have been discovered. Although the protein coding genes can be predicted based on their structural features viz., open reading frame, and features of the upstream and downstream sequences, it is difficult to predict an ncRNA gene only from its sequence information. Because, there is no open reading frame and no sequence similarity, whatsoever so far found, in the characterized genes, in silico analysis has proven to be difficult. With an aim to discover novel ncRNA in higher plants, we are using the in vitro transcription system developed from tobacco cell nuclei. Our efforts have revealed some conserved regulatory elements in Pol III transcribed genes, which are being used to scan genomic data to find new ncRNA candidates. After their amplification by genomic PCR, these candidate genes can be validated for authenticity by using the in vitro transcription system developed in our lab.
Members
Graduate school of Natural Sciences, Professor
Dr. Yasushi Yukawa
Speciality: Plant molecular biology, RNA biology
Projects : Study of transcripution mechanism of higher plants
Profile Ph.D.
Researcher
Ms. Maki Yukawa
Projects: Production of recombinant proteins in chloroplast (Chloroplast Factory Project)
Profile Master of Science
Graduate student(2nd-year doctor's degree)
Ms. Ayaka Ido
Project: Study of light dependent transcription of gene.
Graduate student(1st-year doctor's degree)
Ms. Shuang Li
Project: Study of Arabidopsis lncRNA
Graduate student(2nd-year master's degree)
Mr. Sota Asano
Project: Chloroplast Gene Expression
Graduate student(2nd-year master's degree)
Ms. Na Renhu
Project: Study of rice ncRNA
Graduate student(1st-year master's degree)
Ms. Xueyang Zhao
Project: Study of light dependent transcription of gene.
Graduate student(1st-year master's degree)
Mr. Ailizati Aili
Contact
Institution |
Graduate School of Natural Sciences, Nagoya City University
|
|---|---|
Principal |
Dr. Yasushi Yukawa
|
Address |
Yamanohata, Mizuho, Nagoya 467-8501, Japan
|
E-mail |
yyuk@nsc.nagoya-cu.ac.jp
|

