Introduction (Yukawa Lab., Nagoya City University)
Our group has been involved in developing an understanding of the mechanistic aspects of genes expression during transcription as well as translation in higher plants. Our overall aim is to identify the molecular components involved in these fundamental processes of gene expression and understand their regulation at molecular level. Development of in vitro systems that could transcribe any gene or translate any mRNA with high fidelity was central to achieve the above stated objectives. During the years, we have developed an in vitro transcription system from tobacco nuclei and in vitro translation system from tobacco chloroplasts, which are two major assets of our group and are central to most of our research activities. We hope that by developing a deeper understanding of these fundamental processes of life, our studies would have applications in solving global issues like, food, energy and climate change.
Research
Plant in vitro transcription system
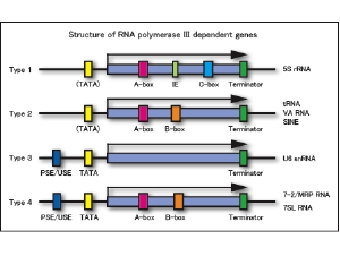
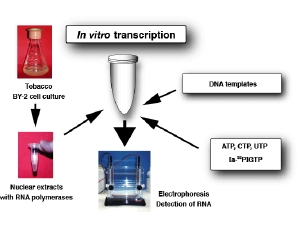 Schematic representation of in vitro transcripiton systemWe are studying various aspects of transcriptional regulation of nuclear genes under cell-free conditions. The in vitro system was developed from BY-2 tobacco cultured cells that are also known as HeLa cells of plants. As eukaryotic cells possess three different RNA polymerases, Pol I, Pol II and Pol III, our in vitro transcription system also supports all these activities. Since our knowledge of Pol III transcription is not as good as for the other two polymerases, our group’s recent efforts are concentrated towards understanding various aspects of Pol III activity. As show below, Pol III transcribed genes (class III genes) in plants have been classified into four types. This classification is different from that of animals and fungi. We therefore believe that a better understanding of plant Pol III based transcription would help in developing a comprehensive understanding of the eukaryotic transcription.
Schematic representation of in vitro transcripiton systemWe are studying various aspects of transcriptional regulation of nuclear genes under cell-free conditions. The in vitro system was developed from BY-2 tobacco cultured cells that are also known as HeLa cells of plants. As eukaryotic cells possess three different RNA polymerases, Pol I, Pol II and Pol III, our in vitro transcription system also supports all these activities. Since our knowledge of Pol III transcription is not as good as for the other two polymerases, our group’s recent efforts are concentrated towards understanding various aspects of Pol III activity. As show below, Pol III transcribed genes (class III genes) in plants have been classified into four types. This classification is different from that of animals and fungi. We therefore believe that a better understanding of plant Pol III based transcription would help in developing a comprehensive understanding of the eukaryotic transcription.
Chloroplast in vitro translation(Chloroplast Factory)
Chloroplast (left) and cytosolic (right) lysates including translation acitivityChloroplasts are plant organelles that are the site for photosynthesis in higher plants. Photosynthesis is a plant-specific biochemical process that produces organic molecules by processing atmospheric CO
2
with the help of energy captured from sun-light. Interestingly, chloroplasts contain a small circular genome that encodes a large number of proteins involved in catalysis of photosynthetic reactions. Since proteins involved in photosynthesis have high turnover rates, chloroplasts require a robust translation system to sustain this high protein turnover. For example, chloroplast translation system is involved in the production of RuBisCO (ribulose-1, 5-bisphosphate carboxylase/oxygenase), which in molar terms is ~50% of all other plastid protein put together and also known to be the most abundant protein on the earth. Chloroplast factory project in our lab focuses on this translational activity and aims to utilize this ability to produce useful proteins in the chloroplast. For this purpose, we have established in vitro translation system from tobacco chloroplast. This system has many advantages, as this system is non-isotopic with high activity and high throughput. Using this system we are trying to study a number of basic aspects related to translation in chloroplasts.
Identification of novel non-coding RNAs from higher plants
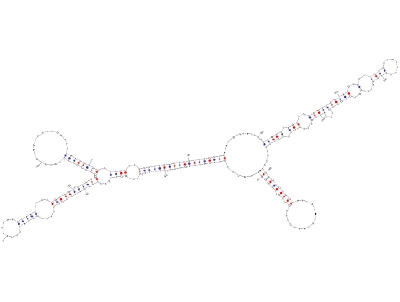 Predicted secondary structure of AtR8 RNAAfter completion of major projects on sequencing of plant genomes, annotation of various structural components of these genomes is in progress. According to the latest estimates the protein coding genes are not so many, however, a large number of non-protein-coding RNA (ncRNA) genes have been discovered. Although the protein coding genes can be predicted based on their structural features viz., open reading frame, and features of the upstream and downstream sequences, it is difficult to predict an ncRNA gene only from its sequence information. Because, there is no open reading frame and no sequence similarity, whatsoever so far found, in the characterized genes, in silico analysis has proven to be difficult. With an aim to discover novel ncRNA in higher plants, we are using the in vitro transcription system developed from tobacco cell nuclei. Our efforts have revealed some conserved regulatory elements in Pol III transcribed genes, which are being used to scan genomic data to find new ncRNA candidates. After their amplification by genomic PCR, these candidate genes can be validated for authenticity by using the in vitro transcription system developed in our lab.
Predicted secondary structure of AtR8 RNAAfter completion of major projects on sequencing of plant genomes, annotation of various structural components of these genomes is in progress. According to the latest estimates the protein coding genes are not so many, however, a large number of non-protein-coding RNA (ncRNA) genes have been discovered. Although the protein coding genes can be predicted based on their structural features viz., open reading frame, and features of the upstream and downstream sequences, it is difficult to predict an ncRNA gene only from its sequence information. Because, there is no open reading frame and no sequence similarity, whatsoever so far found, in the characterized genes, in silico analysis has proven to be difficult. With an aim to discover novel ncRNA in higher plants, we are using the in vitro transcription system developed from tobacco cell nuclei. Our efforts have revealed some conserved regulatory elements in Pol III transcribed genes, which are being used to scan genomic data to find new ncRNA candidates. After their amplification by genomic PCR, these candidate genes can be validated for authenticity by using the in vitro transcription system developed in our lab.
Laboratory members
 Graduate school of Natural Sciences, Professor
Graduate school of Natural Sciences, Professor
Dr. Yasushi Yukawa
Speciality: Plant molecular biology, RNA biology
Projects : Study of transcripution mechanism of higher plants
Profile Ph.D.
![]()

Researcher
Maki Yukawa
Projects: Production of recombinant proteins in chloroplast (Chloroplast Factory Project)
Profile Master of Science
![]()

Graduate student(2nd-year doctor's degree)
Ayaka Ido
Project: Study of light dependent transcription of gene.
Graduate student(2nd-year master's degree)
Toma Fujii
Project: Functional study of plant histone chaperones.
raduate student(2nd-year master's degree)
Sou Li
Project: Functional study of plant long non-coding RNA.
raduate student(1st-year master's degree)
Sota Asano
raduate student(1st-year master's degree)
Na Renhu

Collaborator
Toshiyuki Nagai
Project: rice non-coding RNA.
Publications
![]() Google Scholar Citations (Yasushi Yukawa)
Google Scholar Citations (Yasushi Yukawa)
- 2000
- Yasushi Yukawa, Mamoru Sugita, Nathalie Choisne, Ian Small and Masahiro Sugiura. (2000) The TATA motif, the CAA motif and the poly(T) transcription termination motif are all important for transcription re-initiation on plant tRNA genes. The Plant Journal, 22: 439-447. (
 PMID: 10849359)
PMID: 10849359) - Kazuhito Akama, Volker Junker, Yasushi Yukawa, Masahiro Sugiura and Hildburg Beier. (2000) Splicing of Arabidopsis tRNAMet precursors in tobacco cell and wheat germ extracts. Plant Molecular Biology, 44: 155-165. (
 PMID: 11117259)
PMID: 11117259) - 2001
- Philippe Arnaud, Yasushi Yukawa, Laurence Lavie, Thierry Pelissier, Masahiro Sugiura and Jean-Marc Deragon. (2001) Analysis of the SINE S1 Pol III promoter from Brassica; impact of methylation and influence of external sequences. The Plant Journal, 26: 295-305. (
 PMID: 11439118)
PMID: 11439118) - Yasushi Yukawa, Hao Fan, Kazuo Akama, Hildburg Beier, Hans J. Gross and Masahiro Sugiura. (2001) A tobacco nuclear extract supporting transcription, processing, splicing and modification of plant intron-containing tRNA precursors. The Plant Journal, 28: 583-594. (
 PMID: 11849597)
PMID: 11849597) - 2002
- Olivier Mathieu, Yasushi Yukawa, Masahiro Sugiura, Georges Picard and Sylvette Tourmente. (2002) 5S rRNA genes expression is not inhibited by DNA methylation in Arabidopsis. The Plant Journal, 29: 313-323. (
 PMID: 11844108)
PMID: 11844108) - C. Cloix, S. Tutois, Y. Yukawa, O. Mathieu, C. Cuvillier, M. C. Espagnol, G. Picard and S. Tourmente. (2002) Analysis of the 5S RNA pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature 5S RNA. Genome Research, 12: 132-144. (
 PMID: 11779838)
PMID: 11779838) - Yasushi Yukawa, Jarolav Matousek, Michael Grimm, Lukas Vrba, Gerhart Steger, Masahiro Sugiura and Hildburg Beier. (2002) Plant 7SL RNA and tRNATyr genes with inserted antisense sequences are efficiently expressed in tobacco nuclear extract. Plant Molecular Biology, 50: 713-723. (
 PMID: 12374302)
PMID: 12374302) - Keiko Hasegawa, Yasushi Yukawa, Mamoru Sugita and Masahiro Sugiura. (2002) Organization and transcription of the gene family encoding chlorophyll a/b-binding proteins in Nicotiana sylvestris. Gene, 289: 161-168. (
 PMID: 12036594)
PMID: 12036594) - 2003
- Catherine Cloix, Yasushi Yukawa, Sylvie Tutois, Masahiro Sugiura, Sylvette Tourmente. (2003) In vitro analysis of the sequences required for transcription of the Arabidopsis thaliana 5S rRNA genes. The Plant Journal, 35: 251-261. (
 PMID: 12848829)
PMID: 12848829) - Tadamasa Sasaki, Yasushi Yukawa, Tetsuya Miyamoto, Junich Obokata and Masahiro Sugiura. (2003) Identification of RNA editing sites in chloroplasts transcripts from the maternal and paternal progenitors of tobacco (Nicotiana tabacum): comparative analysis shows the involvement of distinct trans-factors for ndhB editing. Mol. Biol. Evol., 20:1028-1035. (
 PMID: 12716996)
PMID: 12716996) - Keiko Hasegawa, Yasushi Yukawa and Masahiro Sugiura. (2003) In vitro analysis of transcripton initiation and termination from the Lhcb1 gene family in Nicotiana sylvestris: detection of transcription termination sites. The Plant Journal, 33: 1063-1072. (
 PMID: 12631330)
PMID: 12631330) - Keiko Hasegawa, Yasushi Yukawa, Junichi Obokata and Masahiro Sugiura. (2003) A tRNALeu-like sequence within the promoter of an Arabidopsis clock-regulated gene is functional: efficient transcription by an RNA polymerase III-dependent in vitro transcription system. Gene, 307: 133-139. (
 PMID: 12706895)
PMID: 12706895) - Olivier Mathieu, Yasushi Yukawa, Jose-Luis Prieto, Isabelle Vaillant, Masahiro Sugiura and Sylvette Tourmente. (2003) Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana. Nucleic Acids Research, 31: 2424-2433. (
 PMID: 12711688)
PMID: 12711688) - 2004
- Misato Inada, Tadamasa Sasaki, Maki Yukawa, Takahiko Tsudzuki and Masahiro Sugiura. (2004) A systematic search for RNA editing sites in pea chloroplasts: an editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol., 45:1615-1622. (
 PMID: 15574837)
PMID: 15574837) - 2005
- Yasushi Yukawa, Martha Felis, Markus Englert, Michael Stojanov, Jaroslav Matousek, Hildburg Beier and Masahiro Sugiura. (2005) Plant 7SL RNA genes belong to type 4 of RNA polymerase III-dependent genes that are composed of mixed promoters. The Plant Journal, 43: 97-106. (
 PMID: 15960619)
PMID: 15960619) - Maki Yukawa, Takahiko Tsudzuki and Masahiro Sugiura. (2005) The 2005 Version of the Chloroplast DNA Sequence from Tobacco (Nicotiana tabacum) (Updates), Plant Molecular Biology Reporter, 23:1-7. (
 SpringerLink)
SpringerLink) - 2006
- Maki Yukawa, Takahiko Tsudzuki and Masahiro Sugiura. (2006) The chloroplast genome of Nicotiana sylvestris and Nicotiana tomentosiformis: complete sequencing confirms that the Nicotiana sylvestris progenitor is the maternal genome donor of Nicotiana tabacum. Mol Genet. Genomics, 275:367-373. (
 PMID: 16435119)
PMID: 16435119) - Giorgio Dieci, Yasushi Yukawa, Mircko Alzapiedi, Elisa Guffanti, Roberto Ferrari, Masahiro Sugiura and Simone Ottonello. (2006) Distinct modes of TATA box utilization by the RNA polymerase III transcription machineries from budding yeast and higher plants. Gene, 379:12-25. (
 PMID: 16839711)
PMID: 16839711) - Tadamasa Sasaki, Yasushi Yukawa, Tatsuya Wakasugi, Kyoji Yamada and Masahiro Sugiura. (2006) A simple in vitro RNA editing assay for chloroplast transcripts using fluorescent dideoxynucleotides: Distinct types of sequence elements required for editing of ndh transcripts. The Plant Journal, 47:802-810. (
 PMID: 16856984)
PMID: 16856984) - 2007
- Maki Yukawa, Hiroshi Kuroda and Masahiro Sugiura. (2007) A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: effect of pre-mRNA processing on translation in vitro. The Plant Journal, 49: 367-376. (
 PMID: 17156414)
PMID: 17156414) - Yasushi Yukawa, Takaharu Mizutani, Kazuhito Akama and Masahiro Sugiura. (2007) A survey of expressed tRNA genes in the chromosome I of Arabidopsis using an RNA polymerase III-dependent in vitro transcription system. Gene, 392: 7-13. (
 PMID: 17157999)
PMID: 17157999) - Wojchech Plader, Yasushi Yukawa, Masahiro Sugiura and Stefan Malepszy. (2007) The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome: Its composition and comparative analysis. Cellular & Molecular Biology Letters, 12: 584-594. (
 PMID: 17607527)
PMID: 17607527) - Hiroshi Kuroda, Haruka Suzuki, Takahiro Kusumegi, Tetsuro Hirose, Yasushi Yukawa and Masahiro Sugiura. (2007) Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplast. Plant Cell Physiology, 48: 1374-1378. (
 PMID: 17664183)
PMID: 17664183) - 2008
- Maki Yukawa and Masahiro Sugiura. (2008) Termination codon-dependent translation of partially overlapping ndhC-ndhK transcripts in chloroplasts. Proc. Natl. Acad. Sci. USA. 105: 19550-19554. (
 PMID: 19033452)
PMID: 19033452) - 2011
- Yasushi Yukawa, Giorgio Dieci, Mircko Alzapiedi, Asako Hiraga, Katsuaki Hirai, Yoshiharu Yamamoto and Masahiro Sugiura.(2011) A common sequence motif involved in selection of transcription start sites of Arabidopsis and budding yeast tRNA genes. Genomics, 97:166-172. (
 PMID: 21147216)
PMID: 21147216) - Haruka Suzuki, Hiroshi Kuroda, Yasushi Yukawa and Masahiro Sugiura. (2011) The downstream atpE cistron is efficiently translated via its own cis-element in partially overlapping atpB-atpE dicistronic mRNAs in chloroplasts. Nucleic Acids Research, 39: 9405-9412. (
 PMID: 21846772)
PMID: 21846772) - 2012
- Yuka Adachi, Hiroshi Kuroda, Yasushi Yukawa and Masahiro Sugiura. (2012) Translation of partially overlapping psbD-psbC mRNAs in chloroplasts: the role of 5'-processing and translational coupling. Nucleic Acids Research, 40: 3152-3158. (
 PMID: 22156163)
PMID: 22156163) - Yuhta Nomura, Taito Takabayashi, Hiroshi Kuroda, Yasushi Yukawa, Kwanchanok Sattasuk, Mitsuru Akita, Akira Nozawa and Yuzuru Tozawa. (2012) ppGpp inhibits peptide elongation cycle of chloroplast translation system in vitro. Plant Mol. Biol., 78:185-196. (
 PMID: 22108865)
PMID: 22108865) - Juan Wu, Toshihiro Okada, Toru Fukushima, Takahiko Tsudzuki, Masahiro Sugiura and Yasushi Yukawa. (2012) A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Biology, 9: 302-313. (
 PMID: 22336715)
PMID: 22336715) - Elodie Layat, Sylviane Cotterell, Isabelle Vaillant, Yasushi Yukawa, Sylvie Tutois, Sylvette Tourmente. (2012) Transcript levels, alternative splicing and proteolytic cleavage of TFIIIA control 5S rRNA accumulation during Arabidopsis thaliana development. The Plant Journal, 71: 35-44. (
 PMID: 22353599)
PMID: 22353599) - 2013
- Yasushi Yukawa, Kazuhito Akama, Kanta Noguchi, Masaaki Komiya and Masahiro Sugiura. (2013) The context of transcription start site regions is crucial for transcription of a plant tRNALys(UUU) gene group both in vitro and in vivo. Gene, 512: 286-293. (
 PMID: 23103832)
PMID: 23103832) - Maki Yukawa, Masahiro Sugiura. (2013) Additional pathway to translate the downstream ndhK cistron in partially overlapping ndhC-ndhK mRNAs in chloroplasts. Proc. Natl. Acad. Sci. USA. Epub ahead of print. (
 PMID: 23509265)
PMID: 23509265) - 2016
- Ayaka Ido, Shinya Iwata, Yuka Iwata, Hisako Igarashi, Takahiro Hamada, Seiji Sonobe, Masahiro Sugiura and Yasushi Yukawa. (2015) Arabidopsis Pol II-dependent in vitro transcription system reveals role of chromatin for light-inducible rbcS gene transcription. Plant Physiology,170: 642-652.
 PMID: 26662274
PMID: 26662274
Contact
Institution |
Graduate School of Natural Sciences, Nagoya City University
|
|---|---|
Principal |
Dr. Yasushi Yukawa
|
Address |
Yamanohata, Mizuho, Nagoya 467-8501, Japan
|
E-mail |
yyuk@nsc.nagoya-cu.ac.jp
|

Links
- Affiliation
 Nagoya City University
Nagoya City University Graduate School of Natural Sciences, Nagoya City University
Graduate School of Natural Sciences, Nagoya City University- Collaboration (Domestic)
 Dr. Masahiro Sugiura, Nagoya University, Japan
Dr. Masahiro Sugiura, Nagoya University, Japan Dr. Jun-ichi Obokata, Kyoto Prefectural University, Japan
Dr. Jun-ichi Obokata, Kyoto Prefectural University, Japan Dr. Kazuhito Akama, Shimane University, Japan
Dr. Kazuhito Akama, Shimane University, Japan Dr. Yuzuru Tozawa, Ehime University, Japan
Dr. Yuzuru Tozawa, Ehime University, Japan Dr. Seiji Sonobe, University of Hyogo School Science, Japan and Dr. Hisako Igarashi, National Institute for Basic Biology
Dr. Seiji Sonobe, University of Hyogo School Science, Japan and Dr. Hisako Igarashi, National Institute for Basic Biology Dr. Munetaka Sugiyama, University of Tokyo, Japan and
Dr. Munetaka Sugiyama, University of Tokyo, Japan and Dr. Misato Ohtani, RIKEN, Japan
Dr. Misato Ohtani, RIKEN, Japan- Collaboration (Abroad)
 Dr. Sanjay Kapoor, Delhi University South Campus, India
Dr. Sanjay Kapoor, Delhi University South Campus, India Dr. Hildburg Beier, Wuerzburg University, Germany
Dr. Hildburg Beier, Wuerzburg University, Germany Dr. Simone Ottonello,
Dr. Simone Ottonello, Dr. Giorgio Dieci, Parma University, Italy
Dr. Giorgio Dieci, Parma University, Italy